Finalist for best poster award at MIDL 2024 in Paris
Just back from Medical Imaging with Deep Learning (MIDL 2024) in Paris and, again, another succesful conference!
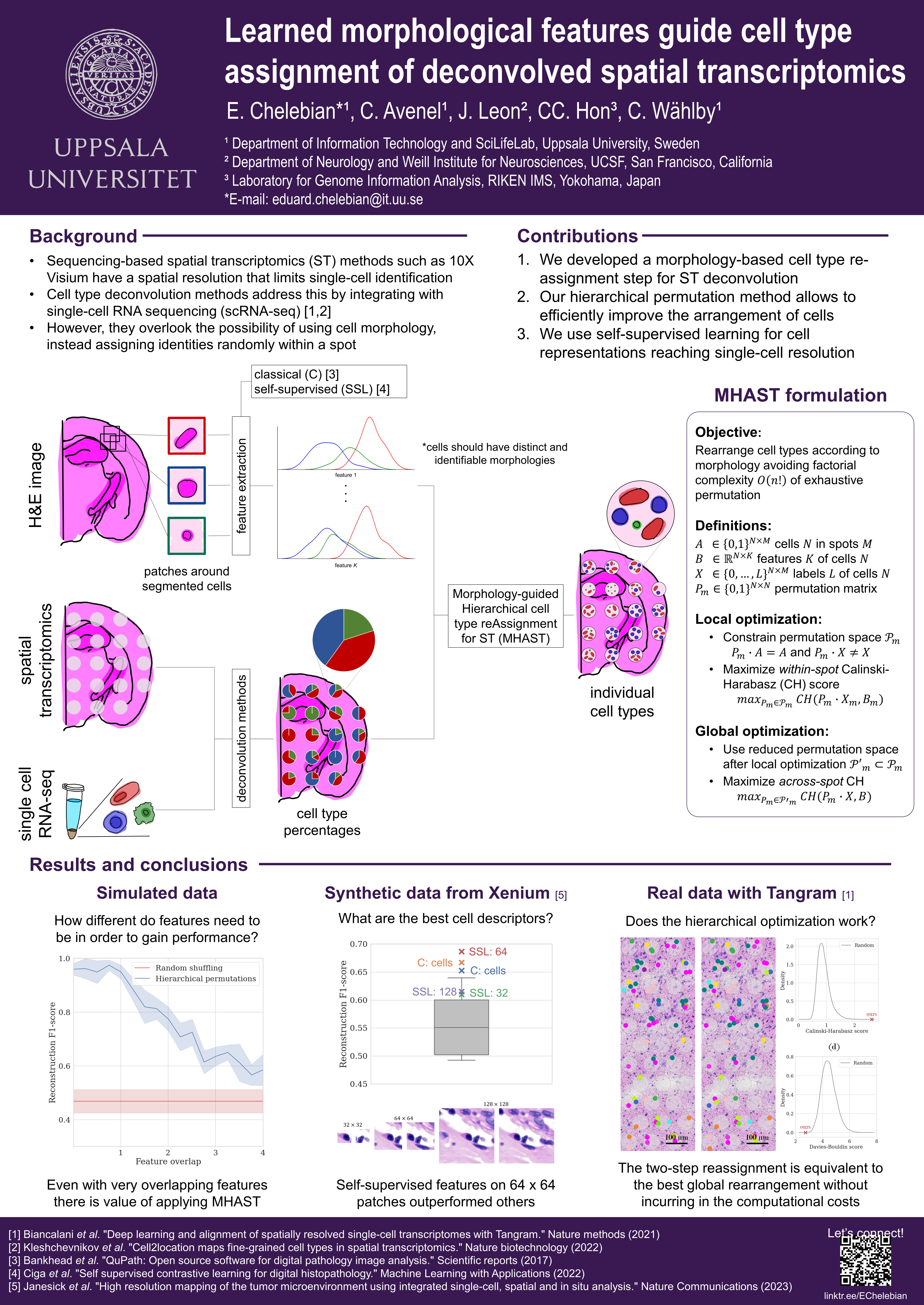
I presented a method we developed during my research visit at RIKEN Yokohama, Japan, entitled “Learned morphological features guide cell type assignment of deconvolved spatial transcriptomics”. The Morphology-guided Hierarchical cell type reAssignment for Spatial Transcriptomics (MHAST) integrates H&E morphology with spatial transcriptomics and single-cell sequencing to increase the resolution of the cell typing.
The poster made was a finalist for the Best Poster Award, recognizing “the highest quality full-length paper presented at the conference” and focusing on “novel methodological concepts with great potential of medical impact”.